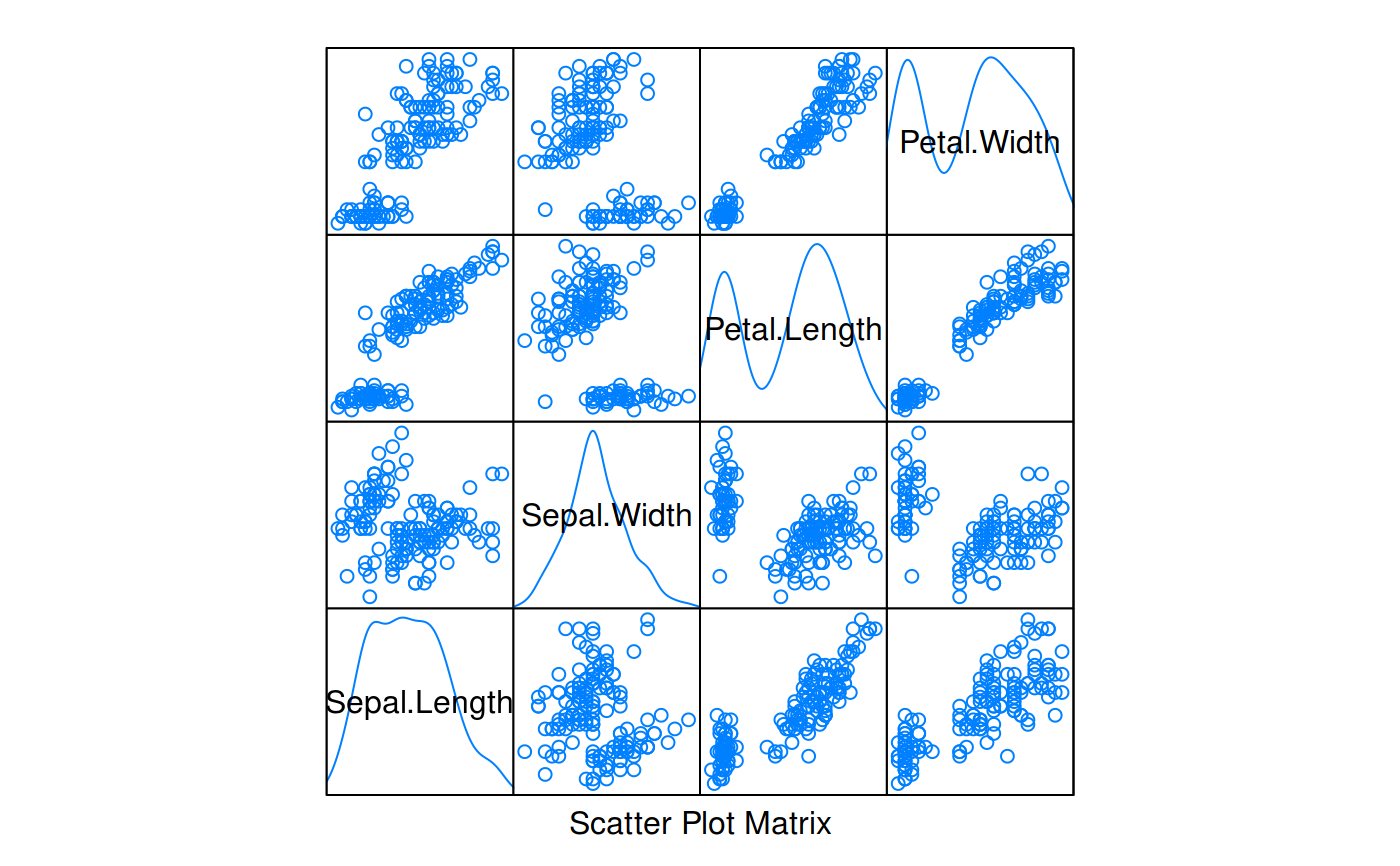
Plots univariate density estimates estimates to be used in a
lattice::splom() call with the diag.panel argument.
Usage
diag.panel.splom.density(
x,
bw = "nrd0",
adjust = 1,
kernel = "gaussian",
weights = NULL,
n = 512,
...
)Arguments
- x
data vector corresponding to that row / column (which will be the same for diagonal 'panels').
- bw
the smoothing bandwidth to be used. The kernels are scaled such that this is the standard deviation of the smoothing kernel. (Note this differs from the reference books cited below.)
bwcan also be a character string giving a rule to choose the bandwidth. Seebw.nrd.
The default,"nrd0", has remained the default for historical and compatibility reasons, rather than as a general recommendation, where e.g.,"SJ"would rather fit, see also Venables and Ripley (2002).The specified (or computed) value of
bwis multiplied byadjust.- adjust
the bandwidth used is actually
adjust*bw. This makes it easy to specify values like ‘half the default’ bandwidth.- kernel
the smoothing kernel to be used. See
stats::density()for options.- weights
numeric vector of non-negative observation weights, hence of same length as
x. The defaultNULLis equivalent toweights = rep(1/nx, nx)wherenxis the length of (the finite entries of)x[]. Ifna.rm = TRUEand there areNA's inx, they and the corresponding weights are removed before computations. In that case, when the original weights have summed to one, they are re-scaled to keep doing so.Note that weights are not taken into account for automatic bandwidth rules, i.e., when
bwis a string. When the weights are proportional to true countscn,density(x = rep(x, cn))may be used instead ofweights.- n
the number of equally spaced points at which the density is to be estimated. When
n > 512, it is rounded up to a power of 2 during the calculations (asfftis used) and the final result is interpolated byapprox. So it almost always makes sense to specifynas a power of two.- ...
Further arguments passed on to
lattice::diag.panel.splom()andlattice::panel.lines().